Molecular Property Prediction using DeBerta
Theoretical explanation and Full Code Walkthrough
Introduction
Molecular property prediction is a pivotal task in fields like drug discovery and material science, offering insights necessary for designing molecules with desirable characteristics, thereby accelerating the development of new drugs and materials. The SELFIES (Self-Referencing Embedded Strings) notation is a robust and flexible chemical language representation designed to overcome the limitations of traditional string-based notations, such as SMILES, by ensuring that every generated string is a valid molecular structure. This characteristic makes SELFIES particularly suitable for computational models aiming to learn from chemical data without being hindered by invalid or ambiguous representations.
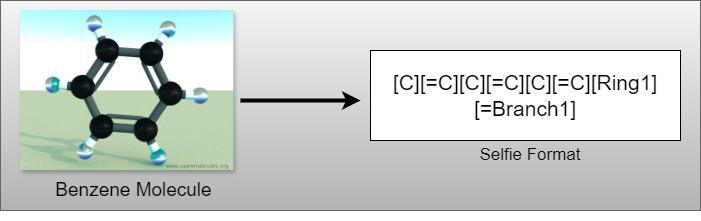
This blog post delves into the challenge of predicting a molecule’s Lipophilicity (Lipo) - a key property influencing its absorption and distribution within the body - using SELFIES notation for input molecules. This prediction task is framed as a regression problem, a critical step towards honing the precision of drug design and material science.
Employing the DeBERTa model, known for its robust transformer architecture, I leverage its capability to capture deep contextual relationships within sequences. The adaptation of DeBERTa for representation learning, in conjunction with SELFIES notation, enables the efficient learning of high-quality molecular representations, enhancing the model’s performance in predicting molecular properties.
Before we explore the intricacies of this approach, let’s gain an understanding of the dataset at our disposal. It consists of a CSV file with 4,200 rows, each representing a molecule in SELFIES notation alongside its numeric Lipo value. This dataset lays the groundwork for our regression model, aimed at predicting molecular properties with precision.
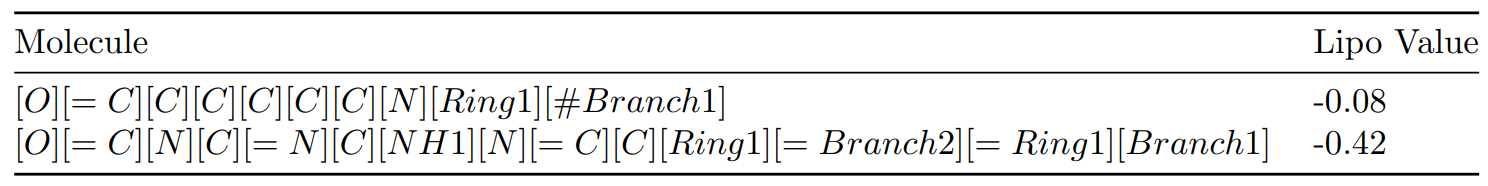
Moreover, we have 100K unlabeled selfies. The idea is to train DeBERTa model with these unlabeled selfies and then finetune the model with labeled dataset for property prediction.
Architecture

The process starts by tokenizing SELFIES strings using a Byte-Level Byte Pair Encoding (BPE) tokenizer, optimized through a large dataset. Next, DeBERTa is pre-trained with these tokens, employing random masking to enhance learning. After pre-training, the model’s architecture is adjusted, replacing the classification head with with regression head. This fine-tuned model, now adept at handling continuous values, targets LIPO property prediction using MSE Loss.
Tokenizer
The initial phase involves creating a tokenizer based on BPE, optimized for the unique structure of SELFIES strings. The objective is to efficiently compress the dataset into a compact vocabulary of subword tokens, formalized as:
\[\min_{V} \sum_{(b_1, b_2) \in V} \log_2(1 + \text{freq}(b_1, b_2))\]Where \(V\) represents the set of byte pairs, and \(\text{freq}(b_1, b_2)\) indicates the frequency of each byte pair in the dataset. This process ensures that the tokenizer captures the molecular structure nuances encoded in SELFIES.
Tokenizer Code
from tokenizers import Tokenizer, Regex
from tokenizers.models import BPE
from tokenizers.pre_tokenizers import Split
from tokenizers.processors import TemplateProcessing
from tokenizers.trainers import BpeTrainer
from transformers import DebertaTokenizerFast
def bpe_tokenizer(path="./data/pretrain.txt"):
# Initialize the tokenizer with the BPE model and unknown token placeholder
tokenizer = Tokenizer(BPE(unk_token="[UNK]"))
# Set pre-tokenizer to split the input on brackets, which are removed from the tokens
tokenizer.pre_tokenizer = Split(pattern=Regex("\[|\]"), behavior="removed")
# Configure post-processor to add special tokens [CLS] and [SEP] around the input
tokenizer.post_processor = TemplateProcessing(single="[CLS] $A [SEP]",
pair="[CLS] $A [SEP] $B:1 [SEP]:1",
special_tokens=[("[CLS]", 1), ("[SEP]", 2)],)
# Define a trainer with special tokens needed for the BPE tokenizer
trainer = BpeTrainer(special_tokens=["[UNK]", "[CLS]", "[SEP]", "[PAD]", "[MASK]"])
# Train the tokenizer on the specified file(s)
tokenizer.train(files=[path], trainer=trainer)
# Save the trained model
tokenizer.model.save("SAVE_DIR")
bpe_tokenizer()
# Convert the BPE into DeBERTa Fast Tokenizer
tokenizer = DebertaTokenizerFast.from_pretrained("./data/bpe")
tokenizer.save_pretrained("./data/debertaTokenizer")
Pre-training with Masking
The pre-training phase for the DeBERTa model leverages the Masked Language Modeling (MLM) technique, a cornerstone of transformer model pre-training. In this approach, a 15% percentage of the input tokens is randomly masked, and the model’s task is to predict these masked tokens based on the context provided by their surrounding, unmasked tokens. The MLM objective is mathematically represented by the loss function:
\[\mathcal{L}_{\text{pretrain}} = -\sum_{\text{masked } i} \log p(x_i | x_{\text{context}})\]This loss function encourages the model to accurately infer the identity of the masked tokens \((x_i)\) using the context provided by the rest of the sequence \((x_{\text{context}})\), thereby gaining a deeper understanding of the molecular structure representations conveyed by SELFIES.
Pre-training code
from transformers import (AutoModelForMaskedLM, DebertaTokenizerFast,
DataCollatorForLanguageModeling,
Trainer, TrainingArguments)
from torch.utils.data import Dataset, DataLoader
# Setup cache directory and model with tokenizer
model = AutoModelForMaskedLM.from_pretrained('microsoft/deberta-base').cuda()
tokenizer = DebertaTokenizerFast.from_pretrained('./data/debertaTokenizer')
# Custom Dataset class to handle SELFIES strings
class LipoPreTrainDataset(Dataset):
def __init__(self, tokenizer, file_path="data/pretrain.txt"):
with open(file_path, 'r') as file:
self.lines = [line.strip() for line in file.readlines()]
self.tokenizer = tokenizer
def __len__(self):
return len(self.lines)
def __getitem__(self, idx):
# Tokenize with truncation, padding, and convert to PyTorch tensors
return self.tokenizer(self.lines[idx], truncation=True, padding='max_length',
max_length=128, return_tensors='pt')
# DataLoader and DataCollator setup for MLM with a 15% masking probability
data_collator = DataCollatorForLanguageModeling(tokenizer=tokenizer, mlm_probability=0.15)
dataset = LipoPreTrainDataset(tokenizer)
# TrainingArguments define training specifics, including epochs, batch size, and output directory
training_args = TrainingArguments(
output_dir='./results',
num_train_epochs=10,
per_device_train_batch_size=16,
save_steps=100,
logging_dir='./logs',
)
# Trainer orchestrates the MLM pre-training, leveraging our dataset and training arguments
trainer = Trainer(model=model, args=training_args, data_collator=data_collator,
train_dataset=dataset)
# Initiates pre-training
trainer.train()
Fine-tuning
After pre-training our model can understand molecular structures through SELFIES representations, the next step involves fine-tuning the pre-trained DeBERTa model on a labeled dataset for predicting LIPO continous values. This step requires modifying the model’s architecture to include a regression head suited for regression tasks. The adaptation to regression is complemented by a shift in the loss function to Mean Squared Error Loss (MSE Loss), defined as:
\[\text{MSE Loss} = \frac{1}{n} \sum_{i=1}^{n} (\hat{y}_i - y_i)^2\]Here, (n) is the number of samples, (\hat{y}_i) is the model’s predicted LIPO value for the (i^{th}) molecule, and (y_i) represents the corresponding actual LIPO value. This phase aims to minimize the differences between the predicted and actual LIPO values, refining the model’s accuracy in property prediction.
Custom Dataset Preparation
We begin by creating a LipoDataset class, which loads our dataset from a CSV file, tokenizes the SELFIES strings, and pairs them with their LIPO values for model training:
from torch.utils.data import Dataset
class LipoDataset(Dataset):
def __init__(self, filepath, tokenizer, max_length=512):
self.data = []
with open(filepath, 'r') as file:
next(file) # Skip header
for line in file:
selfies, prop = line.strip().split(',')[0], line.strip().split(',')[-1]
encoding = tokenizer(selfies, return_tensors='pt', padding='max_length', truncation=True, max_length=max_length)
self.data.append((encoding, float(prop)))
def __len__(self):
return len(self.data)
def __getitem__(self, idx):
encoding, lipo = self.data[idx]
return {**encoding, 'labels': torch.tensor(lipo)}
Model and Training Setup
We adjust the DeBERTa model for sequence classification with a single label to perform regression. The dataset is split into training and test sets, and we define our training loop with MSE loss calculation for regression:
import torch
from transformers import DebertaForSequenceClassification,
DebertaTokenizerFast
from sklearn.model_selection import train_test_split
# Initialize tokenizer and model
tokenizer = DebertaTokenizerFast.from_pretrained('./data/debertaTokenizer')
model = DebertaForSequenceClassification.from_pretrained('./checkpoint', num_labels=1).cuda()
# Prepare dataset
dataset = LipoDataset('data/esol.csv', tokenizer, max_length=128)
train_dataset, test_dataset = train_test_split(dataset, test_size=0.3, random_state=1337)
# Training arguments
training_args = TrainingArguments(
output_dir='./results-finetune',
num_train_epochs=10,
per_device_train_batch_size=16,
save_steps=100,
save_total_limit=1,
overwrite_output_dir=True,
)
# Define a function to compute metrics
def compute_metrics(pred):
labels = pred.label_ids
preds = pred.predictions
mse = mean_squared_error(labels, preds)
return {'mse': mse}
# Trainer for fine-tuning with pretrained weights
trainer = Trainer(
model=model_pretrained,
args=training_args,
train_dataset=train_dataset,
eval_dataset=test_dataset,
tokenizer=tokenizer,
compute_metrics=compute_metrics # Pass the compute_metrics function here
)
trainer.train()
Results
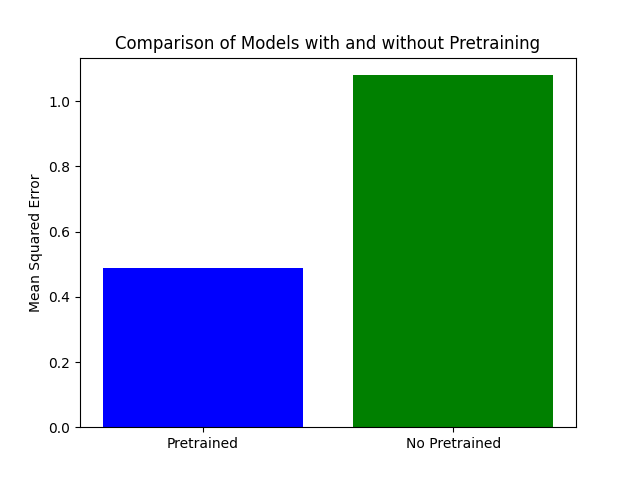
The chart compares the MSE loss on a test dataset for two models: one with pretrained weights and one with random weights. Contrary to expectations, the model with random weights has a higher MSE loss compared to the pretrained model. The pretrained model demonstrates better performance with a significantly lower MSE loss..
Conclusion
In this blog post, you learned how to create a custom tokenizer for a custom dataset. Then we, pre-trained the DeBERTa model and fine-tuned it with a regression task. Moreover, this approach is applicable to multiple domains. Please feel free to reach out to the comment section if you have any doubts.
Reference
- https://selfies.readthedocs.io/en/latest/
- https://huggingface.co/docs/transformers/model_doc/deberta
- Yüksel, Atakan, et al. “SELFormer: Molecular Representation Learning via SELFIES Language Models.” ArXiv, 2023, /abs/2304.04662. Accessed 17 Mar. 2024.
- Fabian, Benedek, et al. “Molecular Representation Learning with Language Models and Domain-relevant Auxiliary Tasks.” ArXiv, 2020, /abs/2011.13230.